Bika Biobank 0.9 Frisky Francolin
|
Cape Town 15 Dec 2016 Frisky Francolin 0.9 complies with year 1 milestones of B3Africa, an European Union Horizon2020 project with four more years to go, and addresses the use cases of the Stellenbosch University Biobank (NSB) at Cape Town's Tygerberg academic hospital. NSB offers biospecimen processing and storage services in collaboration with Tygerberg's pathology departments, SANBI at the University of Western Cape, Rutgers University and the Scripps Research Institute for Regenerative Medicine. Biobanking functionality and workflows added in 2016 include Project and Collection management, Sampling Kit Assembly from inventory components, their Distribution and return, Specimen Preparation, Aliquots, Instrument interfaces, Freezer and Inventory Storage management. The superb work of coders Hocine "The Machine" from SANBI/UWC, and Campbell Mckellar-Basset, must be highlighted. Unbelievable - enjoy the Summer break! Call for participationVersion 0.9 sports use cases and workflow rules that do not necessarily apply everywhere else, or as strictly, was harshly under-resourced and confined by rigid grant milestones. For Bika Biobank to reach release candidate state in 2017, the best road ahead is to optimise the code and re-factor it where necessary. Writing more unit tests and integrating dynamically created user documentation to track the most current functionality, using Sphinx, will go a long way to improve the system's maintainability.
|
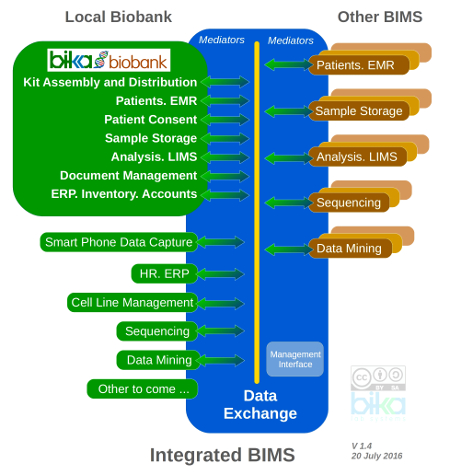
Bika Biobank targets a modular and interoperable biobank management system. Mature OpenHIM is a strong contender as Data Exchange - like Bika it is geared for multiple ontologies, and widely used with strong developer base.
|